
Cells are constantly making decisions that lead to differentiation. For instance, cells in an embryo make a series of decisions that determine whether they will become neurons in some cases and muscle cells in others. How do cells make these decisions?
Researchers at Texas A&M University and North Carolina State University are determining how cells facilitate decision-making processes. Through this work, they hope to precisely measure the concentrations of specific vital signaling proteins within cell tissues. In addition, they will use the measurements to develop mathematical models that can predict and control cellular differentiation.
This study was recently published in ACS Omega.
“We want to understand differentiation decisions, so we can ultimately harness them,” said Dr. Gregory Reeves, an associate professor in the Artie McFerrin Department of Chemical Engineering at Texas A&M. “We are engineering tools to understand cell differentiation and describe the processes through equations. To accomplish these tasks, we need to understand the concentrations of the proteins in live tissues.”
However, determining the concentrations of key signaling proteins can be extremely difficult. To combat this issue, Reeves collaborated with North Carolina State University researchers who used an experimental and analytical framework to develop mix-and-read assays. Mix-and-read assays mean that critical reagents are placed in combination with a lysed cell, allowing for luminescence detection if the target protein is present.
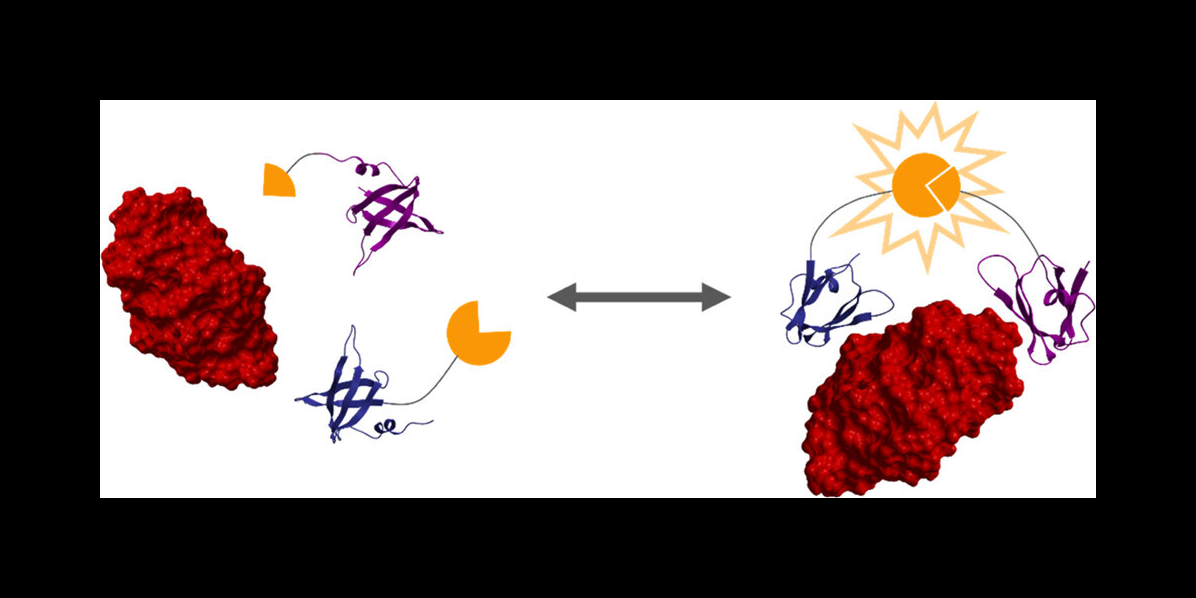
The researchers then used a protein engineering technique to create two proteins that bind strongly to a target protein — in this case, lysozyme. These two protein binders are fused to two halves of luciferase, an enzyme that creates bioluminescence, as you would see in a firefly.
"When the target protein is bound by the two engineered protein binders, it brings the two halves of luciferase together to create bioluminescence, which we can use to take measurements," Reeves said.
Researchers from Reeves’ lab analyzed a mathematical model of this method to predict how much bioluminescence results from the binding events, allowing them to determine the sensitivity of the assay. This, in turn, will help researchers gain a deeper understanding of how and why cells make differentiation decisions.
The broader impacts of this study include using this technique to detect the presence of target proteins, such as antibodies or upregulated cancer markers, in a cellular lysate.
“Other applications, which we will use in my lab, include allowing us to cleanly measure some proteins that were not able to be measured previously in live tissues,” said Reeves.
The researchers also hope to further apply these methods to other classes of molecules that are difficult to detect in live tissues, such as mRNA.
This work is in collaboration with the lead author Nikki McArthur, alongside Dr. Balaji Rao, Dr. Carlos Cruz-Teran and Apoorva Thatavarty from the Department of Chemical and Biomolecular Engineering at North Carolina State University. The research was funded by the Eunice Kennedy Shriver National Institute of Child Health and Human Development, one of the National Institutes of Health.
The research is now funded through the Texas A&M Engineering Experiment Station, a state agency that solves problems through applied research and development, and collaboration with industry, government and academic partners.